BALTIMORE — An international team of scientists has developed a streamlined approach to rapidly sequence the genomes of threatened and endangered species. This breakthrough technology aims to support the ambitious Earth BioGenome Project (EBP) – an effort to catalog the genomes of all of our planet’s biodiversity within the next decade.
Sequencing the genomes of the world’s endangered plants and animals may seem like an odd priority, given more immediate threats like habitat loss and climate change. However, these genetic blueprints could unlock valuable insights to inform conservation strategies, help monitor population health, and even reveal genetic solutions to boost species resilience.
The primary barrier is that genome sequencing remains a major technical hurdle requiring specialized expertise and equipment. Right now, experts using cutting-edge techniques can only assemble a few high-quality vertebrate genomes per year. However, the EBP has set a target to sequence, assemble, and annotate genomes for all of Earth’s estimated 1.8 million cataloged species – including mammals, birds, reptiles, amphibians, fish, insects, plants, and fungi.
To reach this goal, the pace of high-quality genome mapping needs to accelerate by around 100 times within the next seven years. An international team led by scientists from Johns Hopkins University, the Vertebrate Genomes Project (VGP), and the European Reference Genome Atlas (ERGA) have now demonstrated an automated workflow that could provide the essential speed-up.
“Being able to access that genetic information will have huge implications for understanding human health and evolution,” says lead author Michael Schatz, a Bloomberg Distinguished Professor of Computer Science, Biology, and Oncology at Hopkins, in a statement. “A lot of work on drug compounds starts in mice and other animal models, so understanding their genomes and the genomes of other animals directly benefits us.”
The standard approach to assembling high-quality reference genomes involves combining short, accurate DNA reads with long-range sequencing or mapping data. This gives the complete sequence and the structure and organization of the chromosomes.
The team built their new pipeline using Galaxy – an open web-based platform developed for large-scale, reproducible genomics analysis. After systematically testing different analysis options, they optimized a mix of advanced sequencing technologies best suited for automated genome mapping.
Their approach also integrates several quality control checkpoints to catch errors and confirm species identity. For example, a complete mitochondrial genome is assembled early on from the data to validate against catalogs.
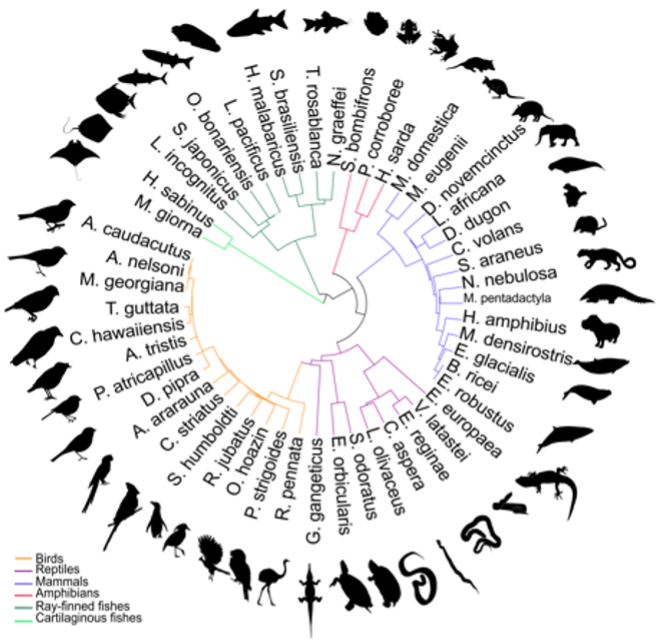
To demonstrate the method’s versatility, the researchers validated genomes across vertebrate species spanning mammals, birds, reptiles, amphibians, and fish. The pipeline improved continuity and genetic resolution even for complex vertebrate genomes over 20 times the size of the human genome.
The Genome Map is Accessible to All Scientists
A key advantage of building the workflow interface through Galaxy is that it makes the analysis accessible to researchers worldwide via web browsers without requiring access to high-powered computing systems. Complete tutorials have also been developed through the Galaxy Training Network portal to teach the approach.
The Galaxy platform allows users of varying skill levels to assemble genomes, whether through a graphical interface or using workflows programmatically. Workflows can also be customized to access distributed high-performance computing systems when large volumes of data need analyzing.
This means that scientists across disciplines and regions – including those studying endangered species – can access top-quality genome sequencing capabilities without costly technology investments. The team hopes this development will empower the wider community to generate foundational genome references and analyze new samples as they are collected over time.
The next phase for the researchers will continue to enhance efficiency for scaling up and keeping pace as sequencing technologies advance. For example, they are working on improvements to allow the integration of emerging ultra-long read nanopore sequencing, which can determine DNA code at unprecedented lengths and even sequence complete chromosomes end-to-end.
There is no doubt that accomplishing the Earth BioGenome Project moonshot will still require extensive coordinated efforts across international working groups, technological fields, policymakers, and funding bodies. However, this latest development demonstrates that the most prohibitive barrier – the actual bioinformatic analytics – is now within reach. The complete library of life’s genomic diversity could become a reality through such collaborative science benefiting conservation.
The study is published in the journal Nature Biotechnology.
